(CS182)Machine Learning Review
Scope of Final Exam
Bayes’ Decision Rule, Discriminant Functions
Maximum Likelihood Estimation, Bayesian Estimation, Paramatric Classification, Model Selection
Paramatric Classification Revisited
Multilayer Perceptrons
Hard-Margin SVM, Soft-Margin SVM, Kernel Extension
PCA, LDA
k-Means Clustering Algorithm, EM algorithm, Use of Clustering
Nonparametric Density Estimation, Nonparametric Classification
CNN, RNN
Voting, Boosting
Cross-Validation, Interval Estimation, Performance Evaluation
Preliminaries
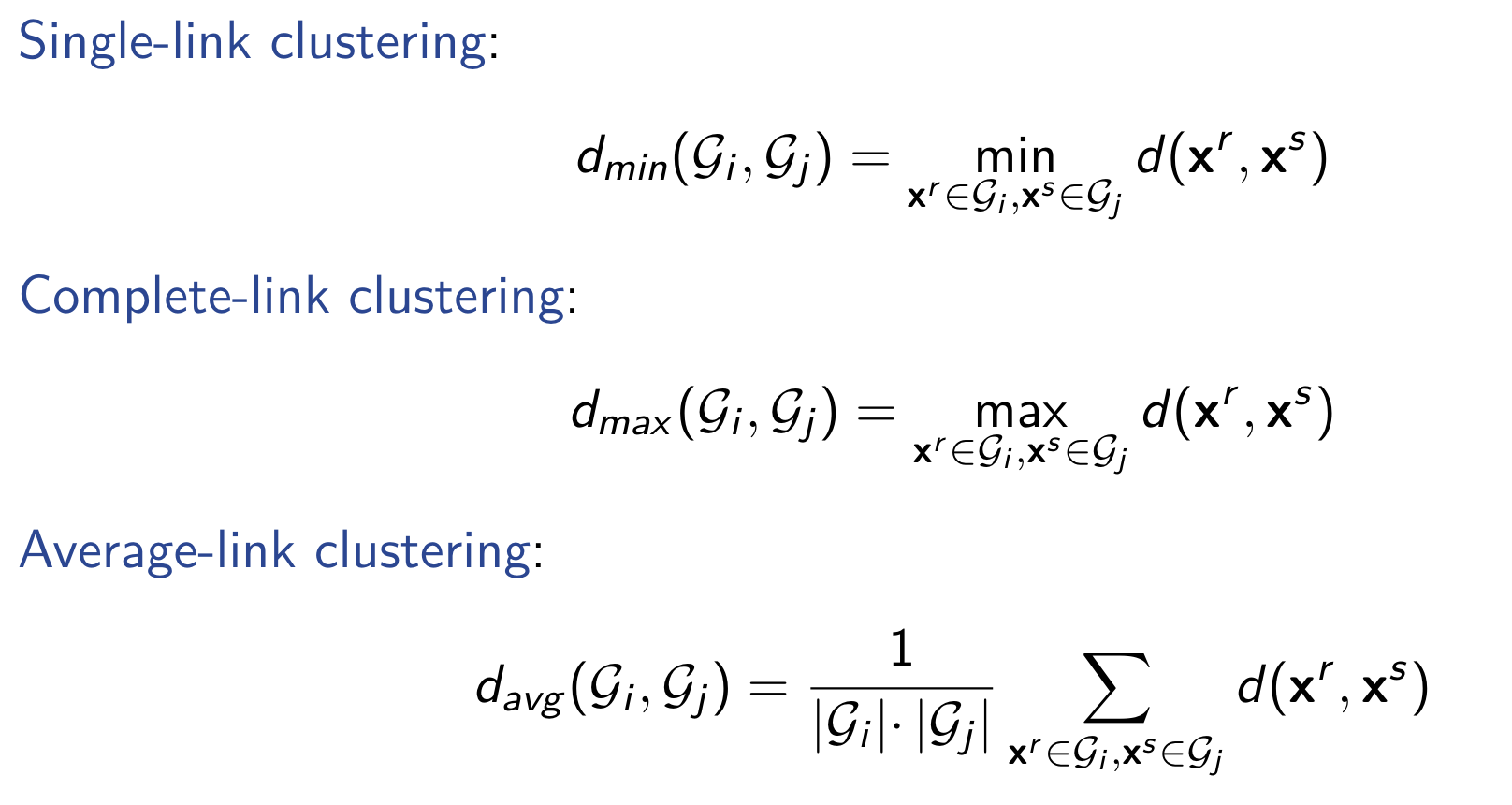
Distance Measures
Distance between instance

Distance between groups
Linear Algebra
Transpose
Matrix
Inverse
Gradient Vector
Positive Semidefinite Matrices
A symmetric matrix
positive semidefinite:
positive definite:
indefinite: both
If
A positive definite matrix can be “factored” as
Eigendecomposition(Spectral Decomposition)
where Q is a square
If
Singular Value Decomposition
If
where
of both
where
their diagonal and 0 elsewhere.
Optimization Primer
Lagrange multipliers can transfer a problem with
Standard foem problem:
Lagrangian
Assume we do not consider the constraints and obtain the optimal value in point
For the first situation, the constrains is ignored and satiesfied automatically. For the second situation, we can obtain the answer by simply calculate the Lagrangian. For the third situation, we should discard the answer. And in conclusion, if
And we have
Above all, we can conclude the Karush-Kuhn-Tucker condition: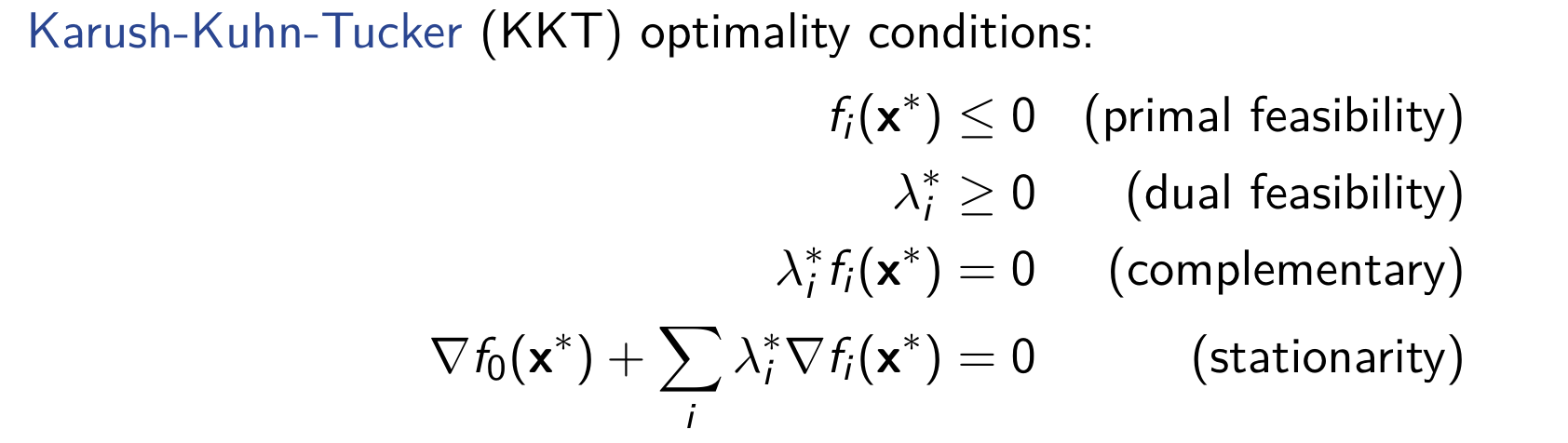
Bayesian Desicion Model
Parameter Estimation
Linear Discrimination
Multilayer Perceptions
Support Vector Machines
Hard-Margin Support Vector Machine
Definition of hyperplane
Definition of distance
If the sample point can be classified properly, we have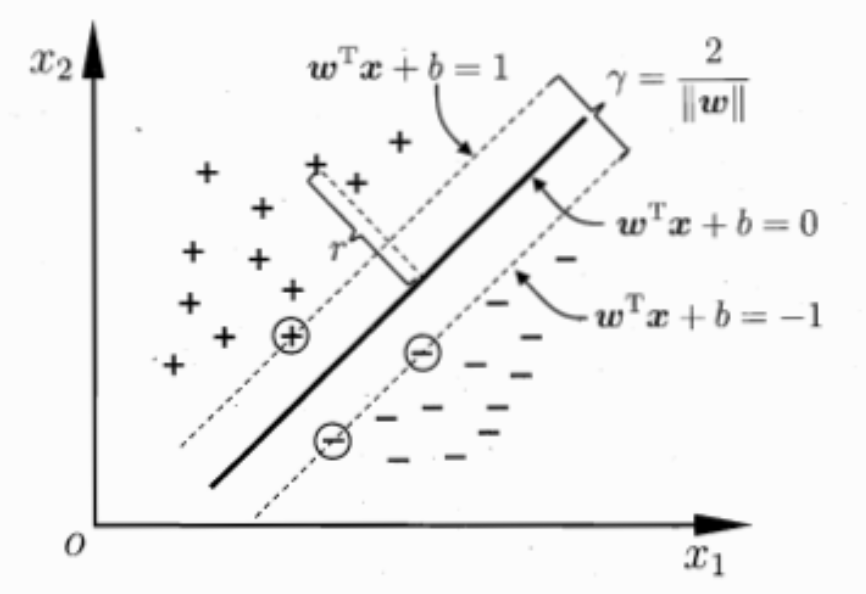
Nearest points to the hyperplane are called support vector, and the sum of distance of two support vectors to the hyperplane from different class is called margin
We want to maximize the distance, or equivalently, minimize
Dual Problem
We have its lagrangian form
Calculate its derivative and we have
And we can have its dual form
It should satisfy KKT conditions
And if we have
where its optimal will be
Soft-Margin Support Vector Machine
Relaxed seperation constriants
Primal optimization problem
Its lagrangian can be written as
Dual problem
And its KKT conditions are
kernel extension
Instead of defining a nonlinear model in the original (input) space, the problem is mapped to a new (feature) space by performing a nonlinear transformation using suitably chosen basis functions.
Similarly, we have
Its dual problem is
Since calculating
And we can rewrite the dual problem as
And the answer is
Polynomial kernel:
where ( q ) is the degree.
E.g., when ( q = 2 ) and ( d = 2 ),
which corresponds to the inner product of the basis function
When ( q = 1 ), we have the linear kernel corresponding to the original formulation.
Support Vector Regression

Minimization Problem:
$$
\text{minimize}{\mathbf{w}, w_0} \quad \frac{1}{2} |\mathbf{w}|^2 + C \sum_t \left( |r^t - f(\mathbf{x}^t)| - \epsilon \right)+
$$
Primal optimization problem:
Lagrangian:
$$
\begin{aligned}
\mathcal{L}(\mathbf{w}, w_0, {\xi^+_t}, {\xi^-_t}, {\alpha^+_t}, {\alpha^-_t}, {\mu^+_t}, {\mu^-_t}) = \frac{1}{2} |\mathbf{w}|^2 + C \sum_t (\xi^+_t + \xi^-_t) \
- \sum_t \alpha^+_t \left[ \epsilon + \xi^+_t - r^t + (\mathbf{w}^T \mathbf{x}^t + w_0) \right] \
- \sum_t \alpha^-_t \left[ \epsilon + \xi^-_t + r^t - (\mathbf{w}^T \mathbf{x}^t + w_0) \right] \
- \sum_t (\mu^+_t \xi^+_t + \mu^-_t \xi^-_t)
\end{aligned}
$$
Dual optimization problem:
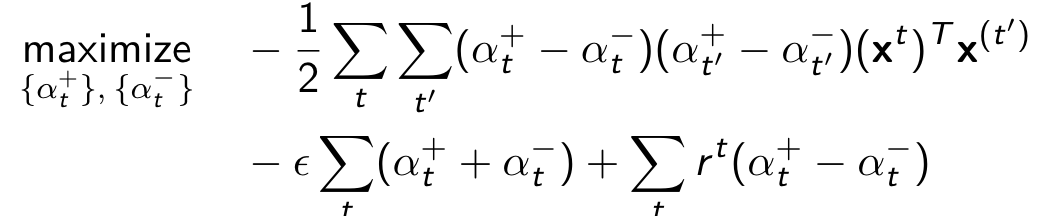
Function Representation:
Dimentionally Reduction
Principal Component Analysis
We aim to fina a linear mapping from d-dimensional input space to k-dimensional with minimum information loss according to some criterion(k<<d)
We have the scalar projection of
we want to find the component that can make the sample points the most spread out, which simply can be achieved by maximize its variance,
And the goal of PCA will be
and we can do eigenvalue decomposition to covairencematrix
And for choosing the detalied number of k, we can have a threshold,i.e.
Factor Analysis
The target of FA is opposite to that of PCA:
PCA (from x to z):
A (from z to x – generative model):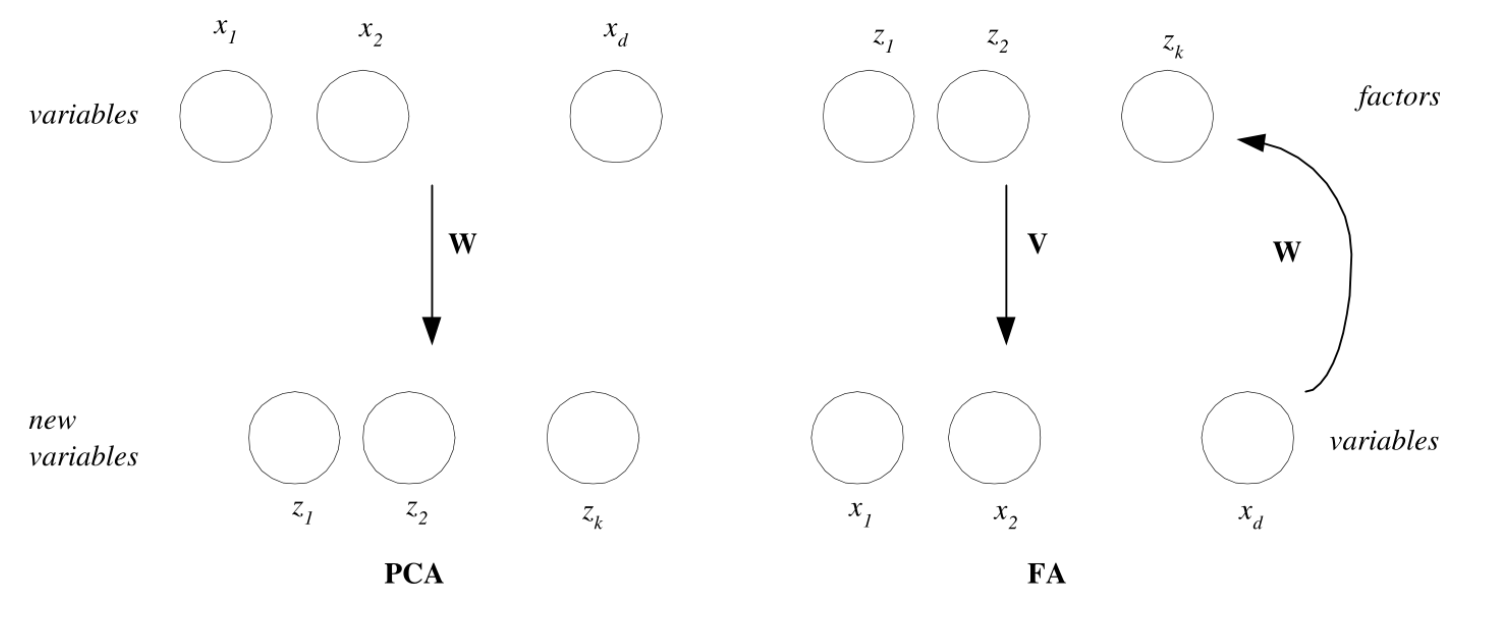
Multiple Dimensional Scaling
We want to embed the points in a lower-dimensional space (e.g., two-dimensional space) such that the pairwise Euclidean distances in this space are as close as possible to those in the original space.
Assuming m sample point in original space have a distance matrix
Let
Centering of data to constrain the solution:
And it is obvious that the sum of the row and column of
Let’s have
$$dist_{i*}^2=\frac{1}{m} \sum_{j=1}^m dist_{ij}^2\quad dist_{j}^2=\frac{1}{m} \sum_{i=1}^m dist_{ij}^2 \quad dist_{i}^2=\frac{1}{m^2} \sum_{i=1}^m \sum_{j=1}^m dist_{ij}^2
And let $\Lambda_{}
Linear Discriminant Analysis
Goal: the classes are well-separated after projecting to a low-dimensional space by
utilizing the label information (output information).
Let
We want to minimize covariance between in-class covariance
Also, we want to maximize the distance between center of classes
$$ ||w^T \mu_0- w^T \mu_1||{2}^{2}
the solution is the eigen vector relevant to
Clustering and Mixture Models
Introduction
Mixture(density) model
Gaussian Mixture Model(GMM)
parameters
K-Means Clustering Algorithm
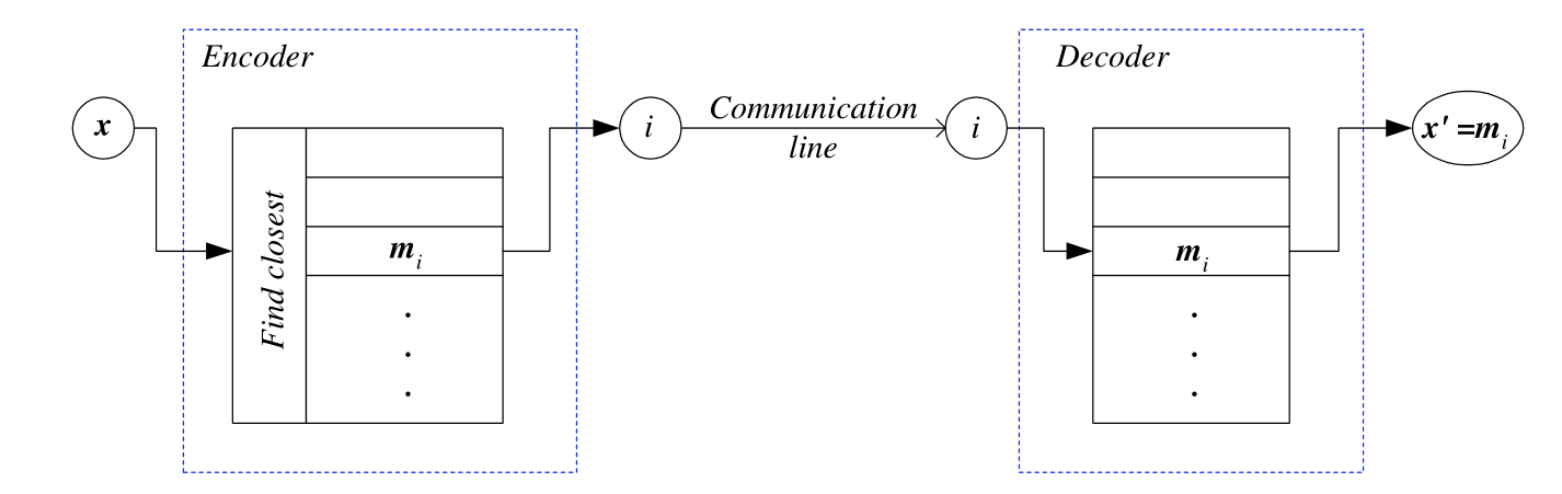
Encoding: from data point
Decoding: from an index
Each data point
Total recontruction error
where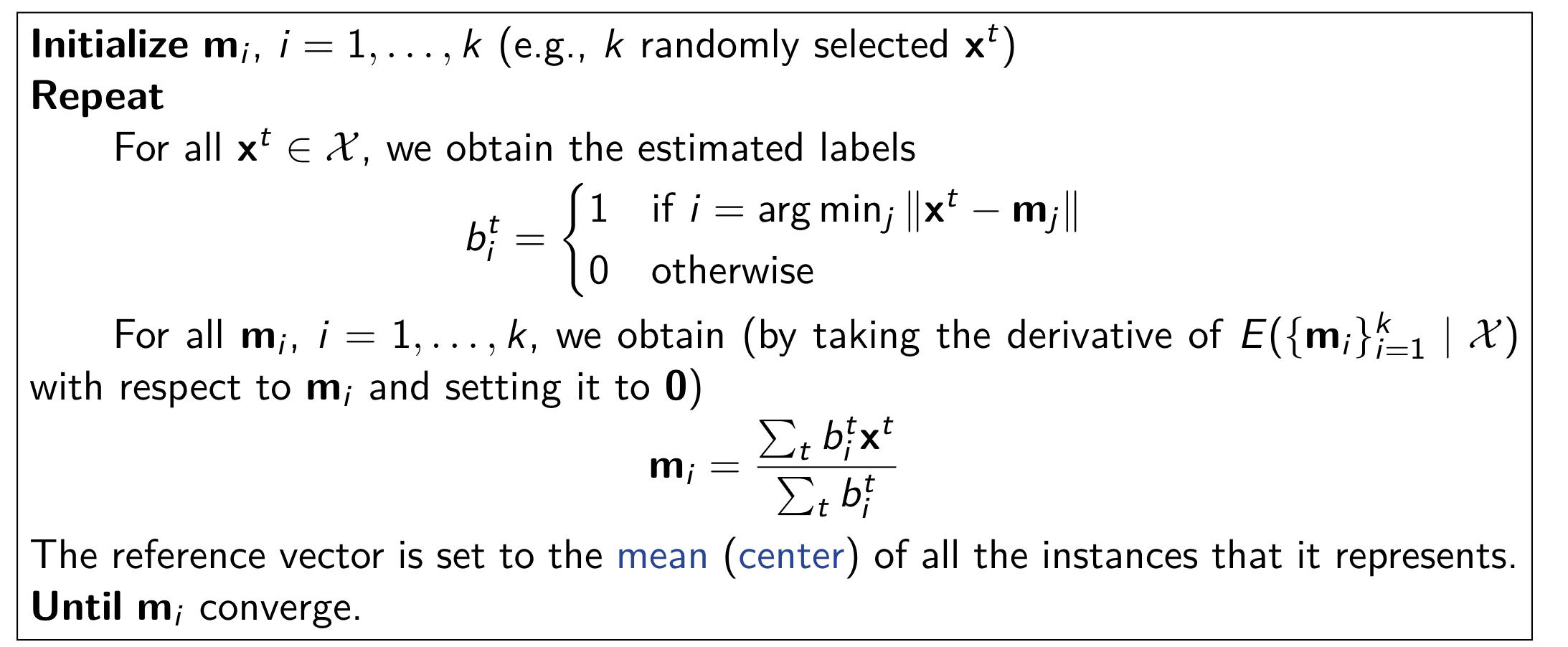
Expectation-Maximization Algorithm
For unobserved data, we can use EM to do estimation. We call this unobserved variables
Let
Since Z is latent variable, it can not be optimized directly, we can calculate the expectation of
E-Step
Infer the distribution of latent variables
M-Step
Find parameter
In one word, EM calculate global optimal by caulate expectation of current parameter and find the parameter that can maximize likelihood expectation.
Use of Clustering
Data Exploration
Like dimensionality reduction methods, clustering can be used for data exploration:
Dimensionality reduction methods: find correlations between features (and thus group features).
Clustering methods: find similarities between instances (and thus group instances).
Clustering allows knowledge extraction through:Number of clusters,Prior probabilities,Cluster parameters
Clustering as Preprocessing
After clustering, the estimated group labels may be seen as the dimensions of a new k-dimensional space.
Mixture of Mixtures in Classification
In classification, when each class is a mixture model composed of a number of
components, the whole density is a mixture of mixtures
withs
Hierarchical Clustering
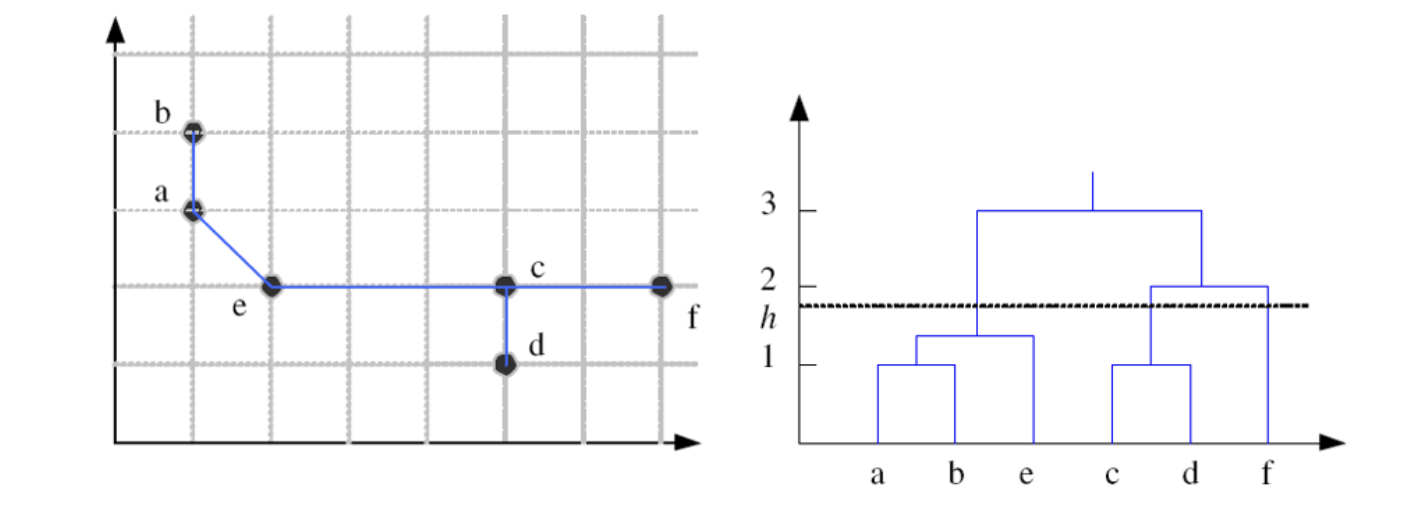
Nonparametric Methods
Deep Learning Models
Deep Autoencoders
An autoencoder (AE) is a feedforward neural network for learning a compressed representation or latent representation (encoding) of the input data by learning to predict the input itself in the output.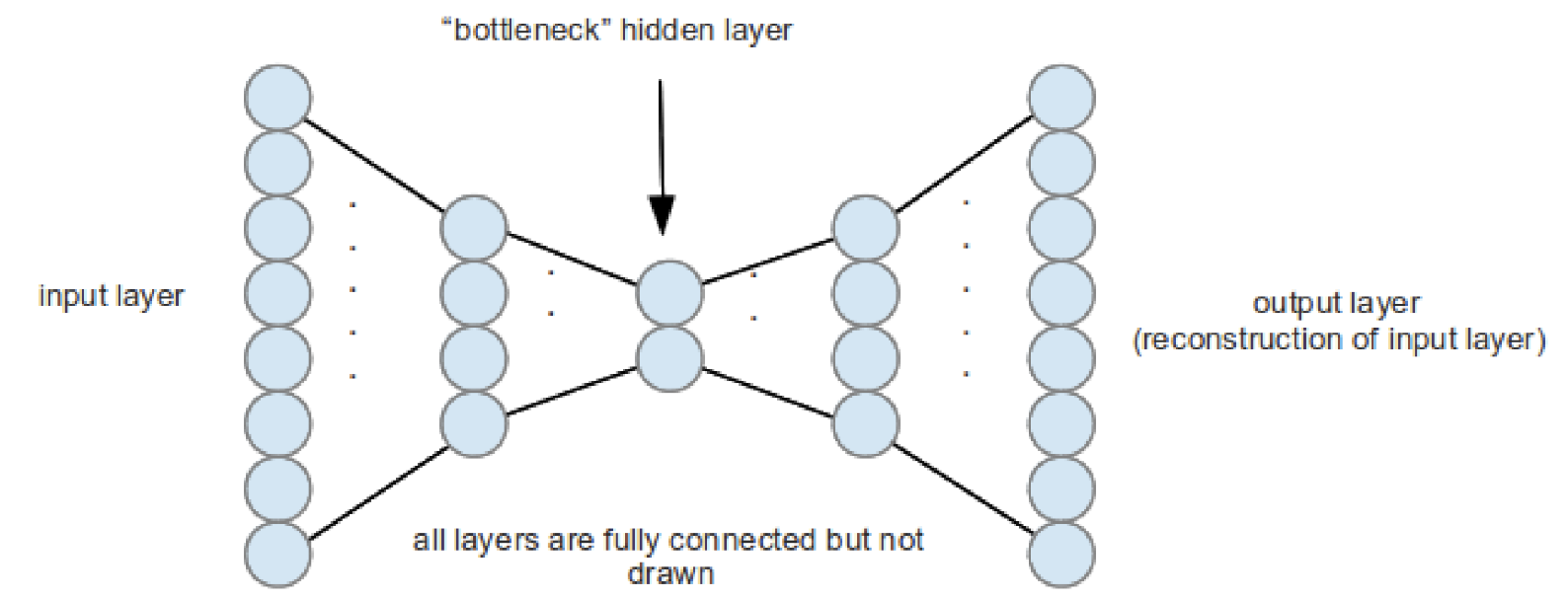
The hidden layer in the middle is constrained to be a narrow bottleneck (with
fewer units than the input and output layers) to ensure that the hidden units
capture the most relevant aspects of the data.
Stacked Denoised Autoendcoder
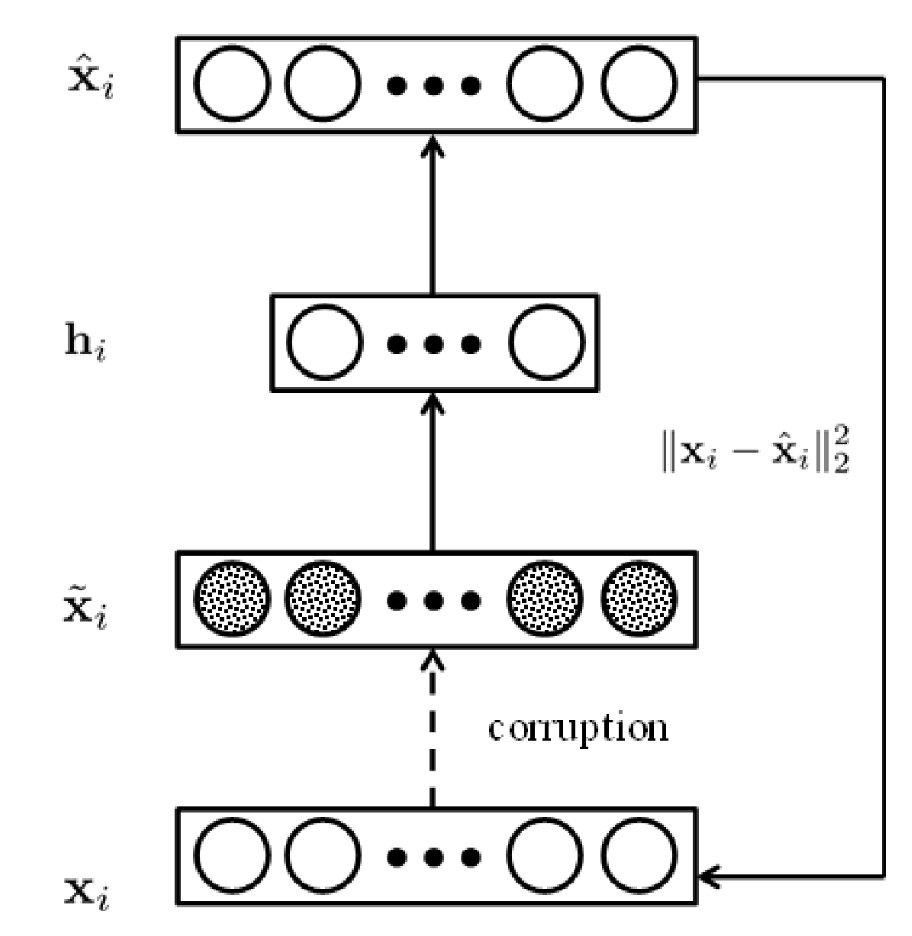
A denoising autoencoder solves the following (regularized) optimization problem:
$$\text{minimize}_{\mathbf{W}1, \mathbf{W}2, \mathbf{w}{01}, \mathbf{w}{02}} \quad \frac{1}{2} \sum_\ell | \mathbf{x}^\ell - \hat{\mathbf{x}}^\ell |_2^2 + \lambda \left( | \mathbf{W}_1 |_F^2 + | \mathbf{W}_2 |_F^2 \right)$$
where
$$\begin{aligned}
\mathbf{h}^\ell &= \sigma(\mathbf{W}1 \tilde{\mathbf{x}}^\ell + \mathbf{w}{01}) \
\hat{\mathbf{x}}^\ell &= \sigma(\mathbf{W}2 \mathbf{h}^\ell + \mathbf{w}{02})
\end{aligned}$$
Here
$$| \mathbf{A} |F^2 = \sum{i,j} a_{ij}^2 \quad \text{where} \quad \mathbf{A} = [a_{ij}] \text{ is a matrix}$$
Stacked denoising autoencoders can be formed by stacking the denoising
autoencoders in a layer-wise manner like deep autoencoders with the same
pretraining, unrolling, and fine-tuning steps.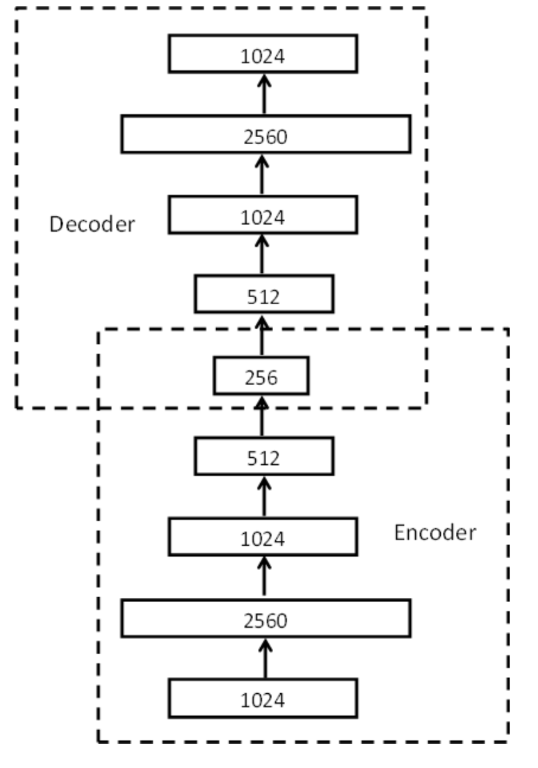
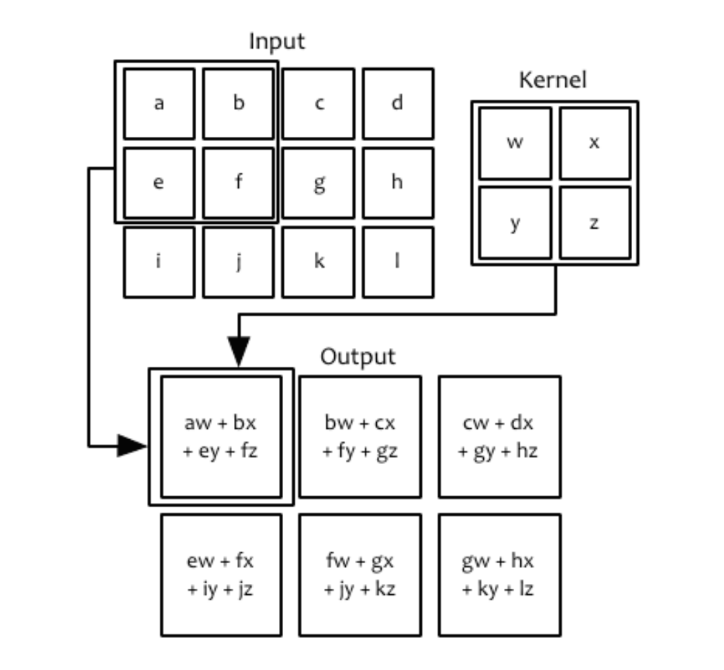
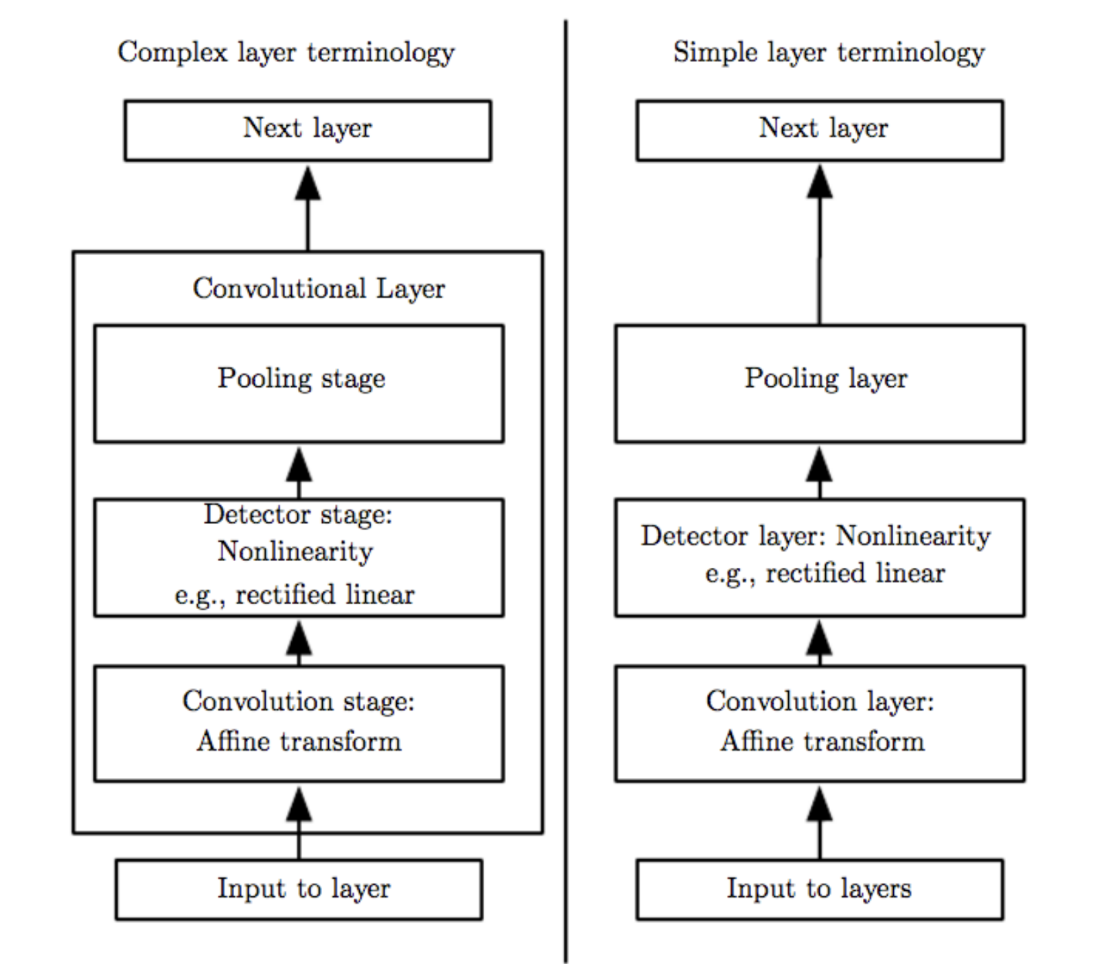
Convolutional Neural Networks
Dropout is very powerful because it effectively trains and evaluates a bagged ensemble of exponentially many deep models which are sub-networks formed by removing (randomly with a dropout rate of say 0.5) units or weights (achieved simply via multiplying their values by zero) from the original CNN.
Recurrent Neural Networks
Recurrent neural networks (RNNs) are extensions of feedforward neural networks for handling sequential data (such as sound, time series (sensor) data, or written natural language) typically involving variable-length input or output sequences.
There is weight sharing in an RNN such that the weights are shared across different instances of the units corresponding to different time steps.
A classical dynamic system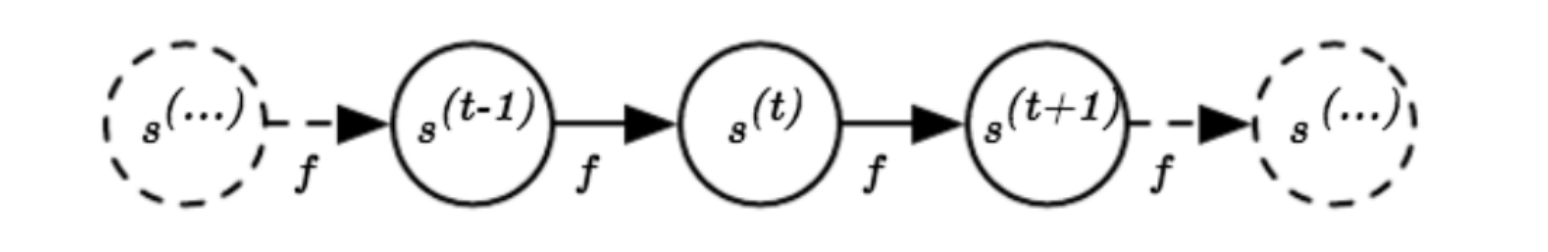
A more generalized form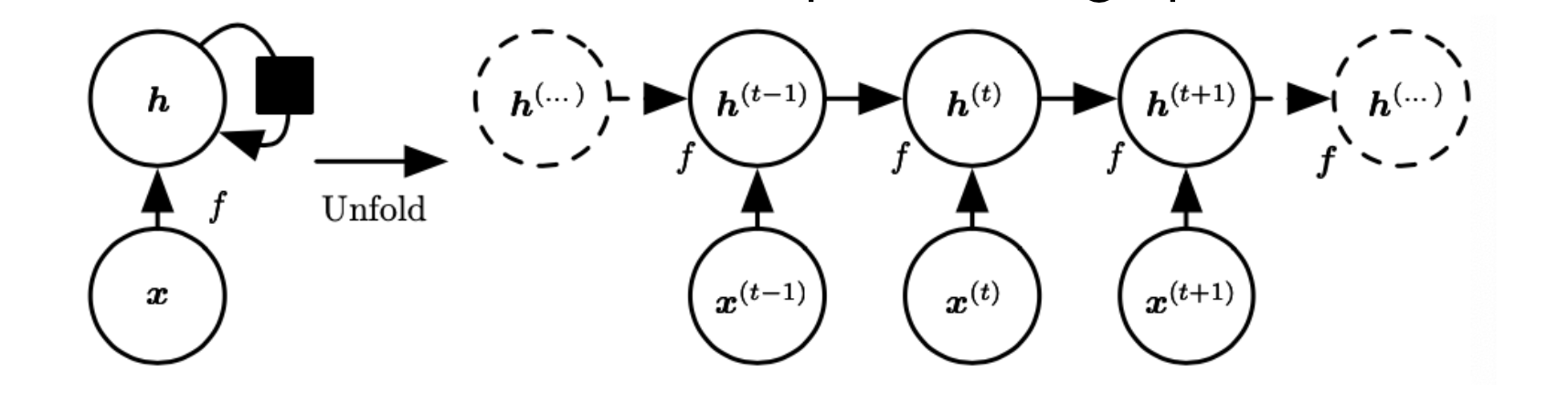
A recurrent neuron with a scalar output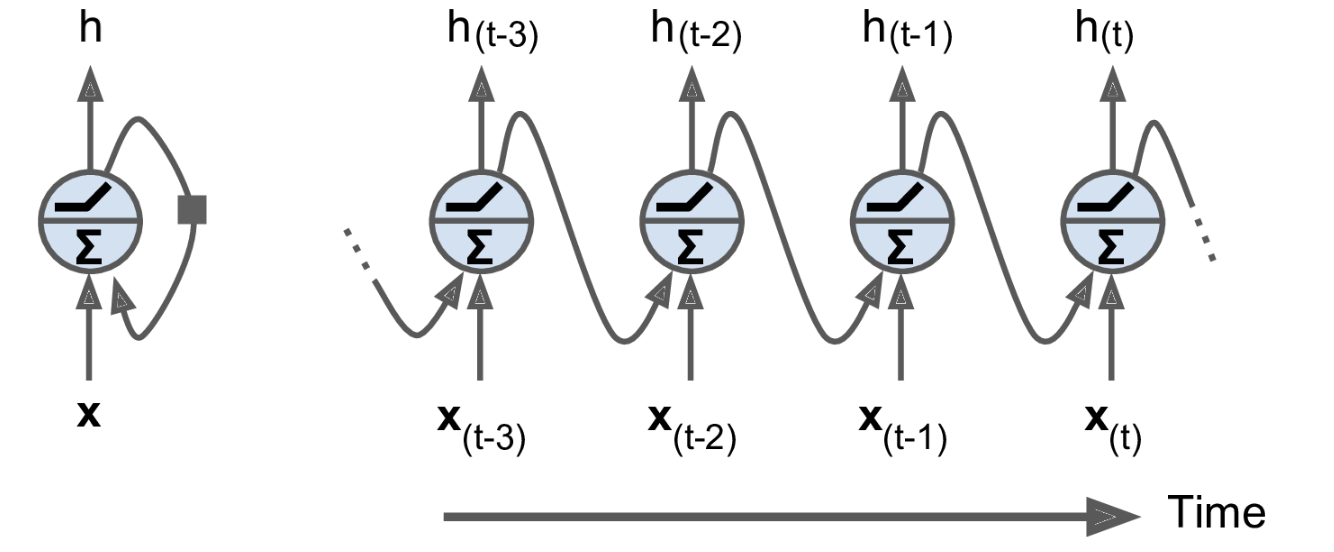
A layer of recurrent neurons with a vector output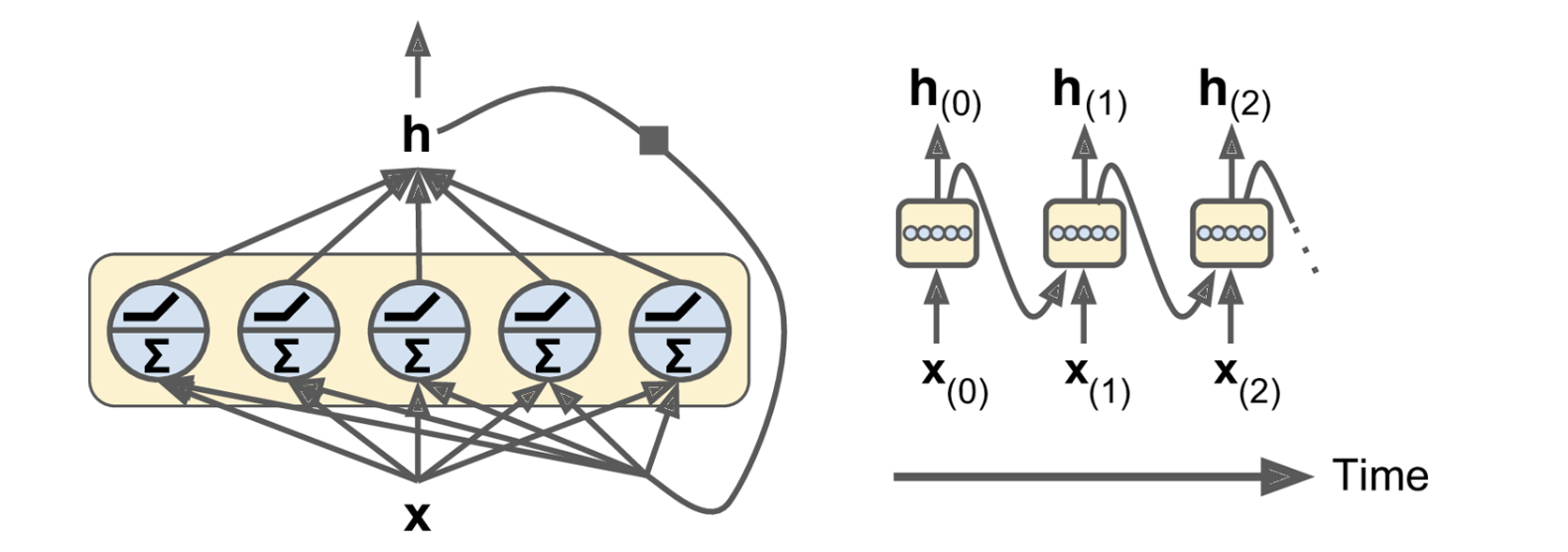
The following gradients (used by gradient-based techniques) can be computed
recursively (details ignored) like the original backpropagation algorithm:
RNN Generalizations
Encoder-Decoder RNN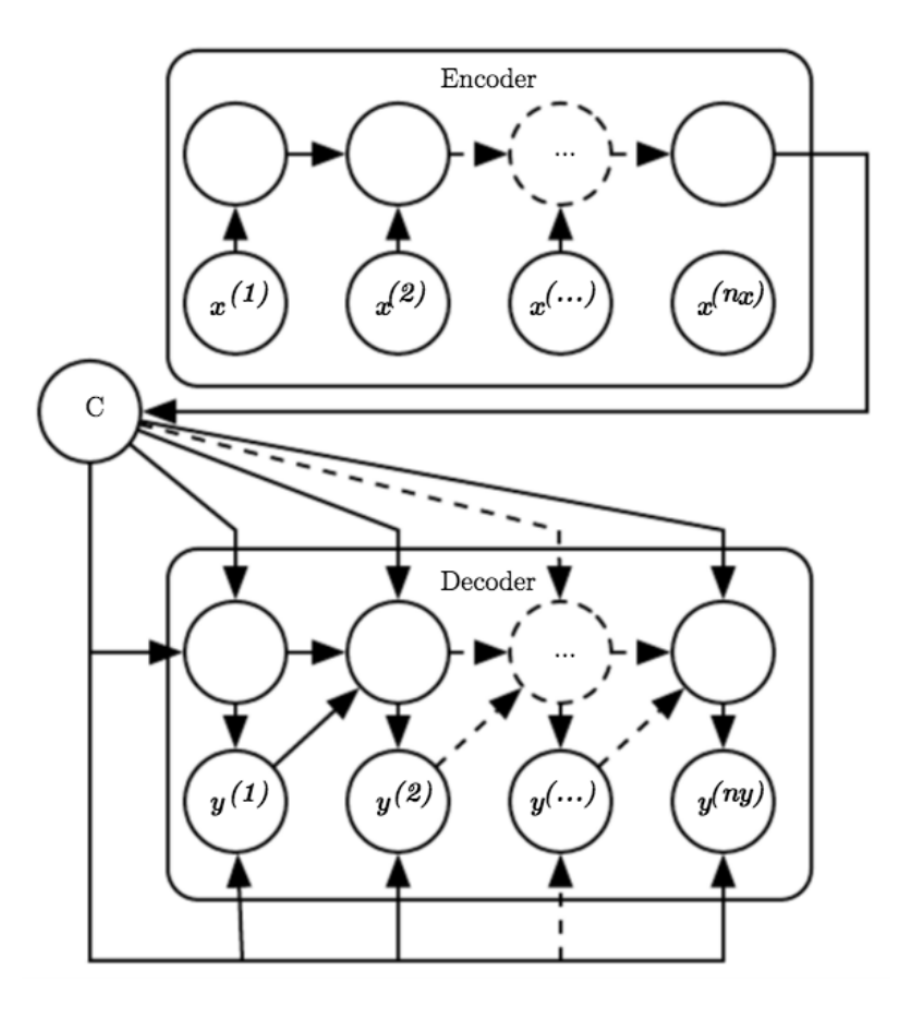
And there are many ways to increase the depth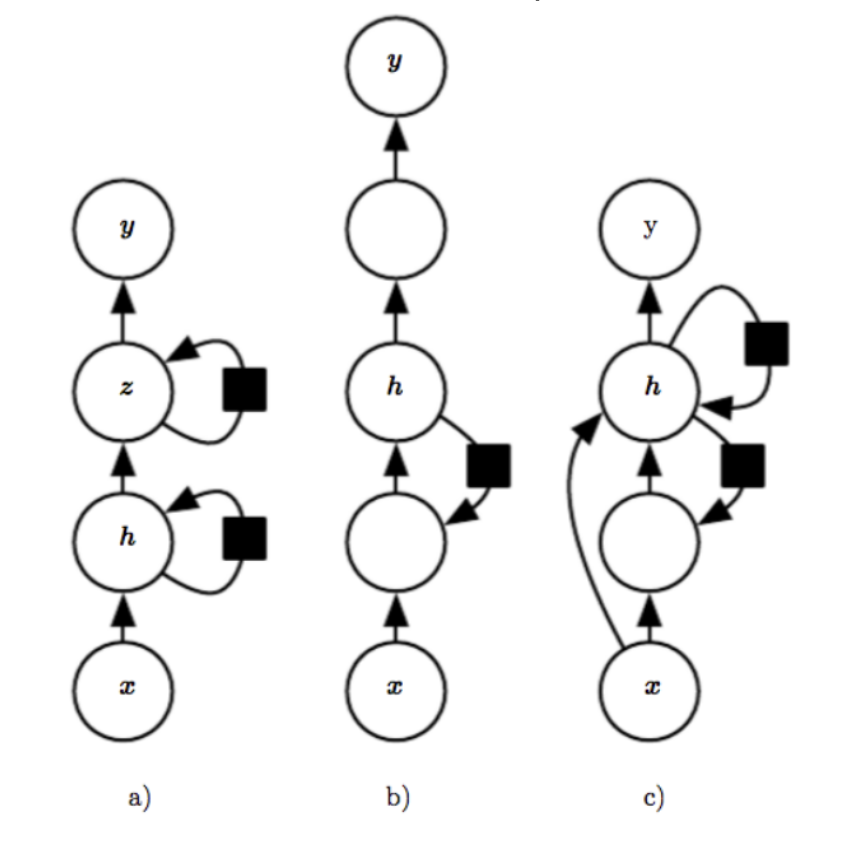
Dependencies between Events in RNNs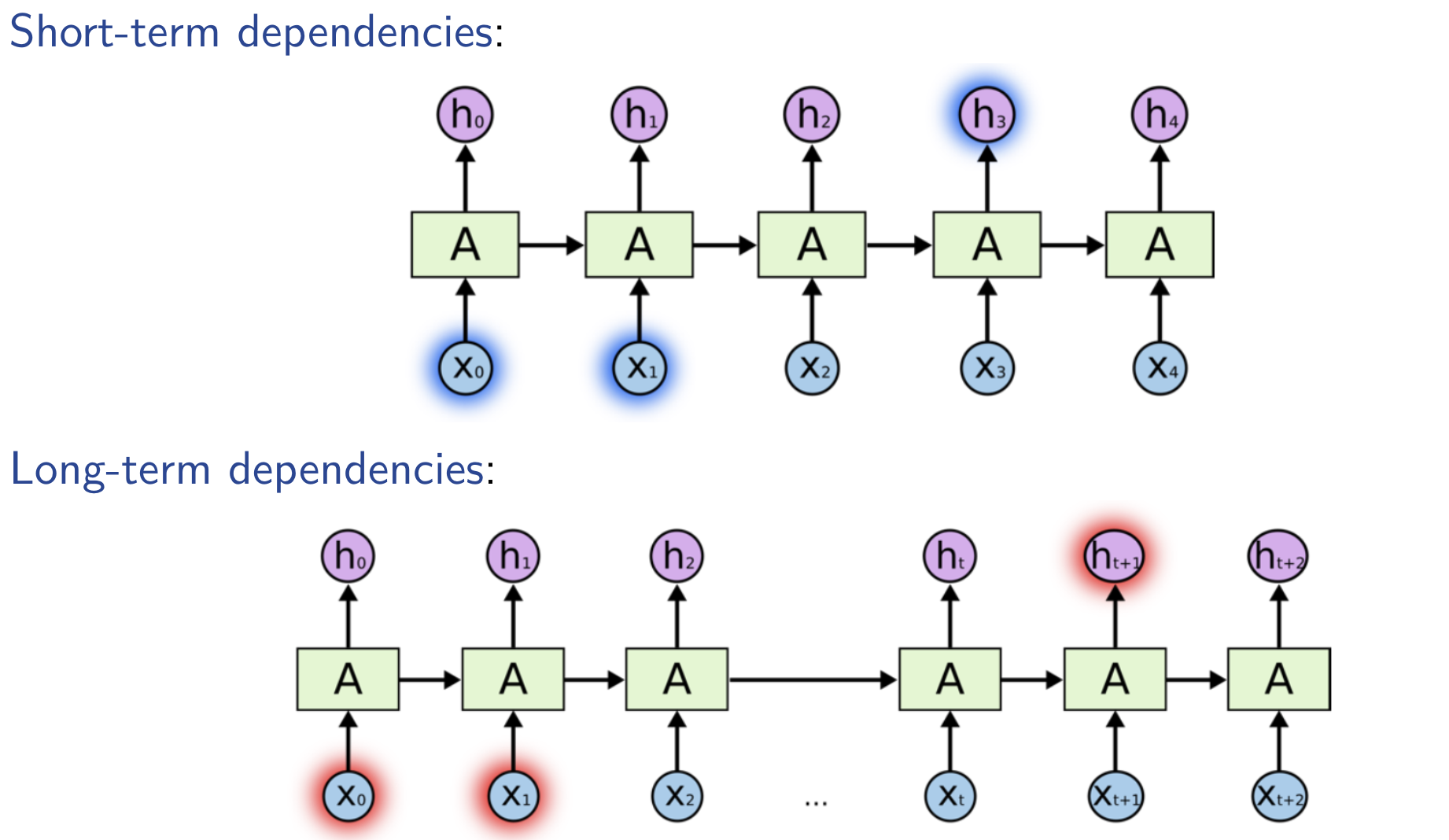
Shortcomings of long-term RNN:1. vanish or explode of gradients.2. computationally intensive
LSTM is based on the idea of leaky units which allow an RNN to accumulateinformation over a longer duration (e.g., we use a leaky unit to accumulate evidence for each subsequence inside a sequence).I Once the information accumulated is used (e.g., suffcient evidence has been accumulated for a subsequence), we need a mechanism to forget the old state by setting it to zero and starting to count again from scratch.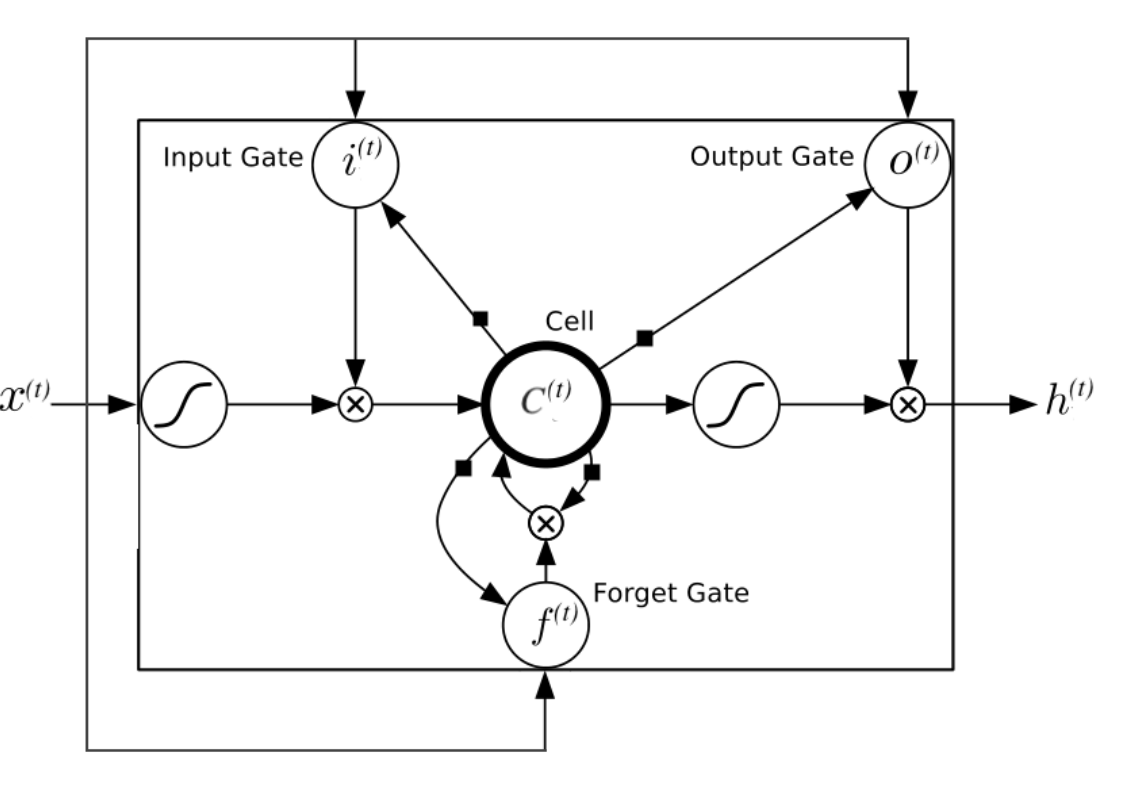
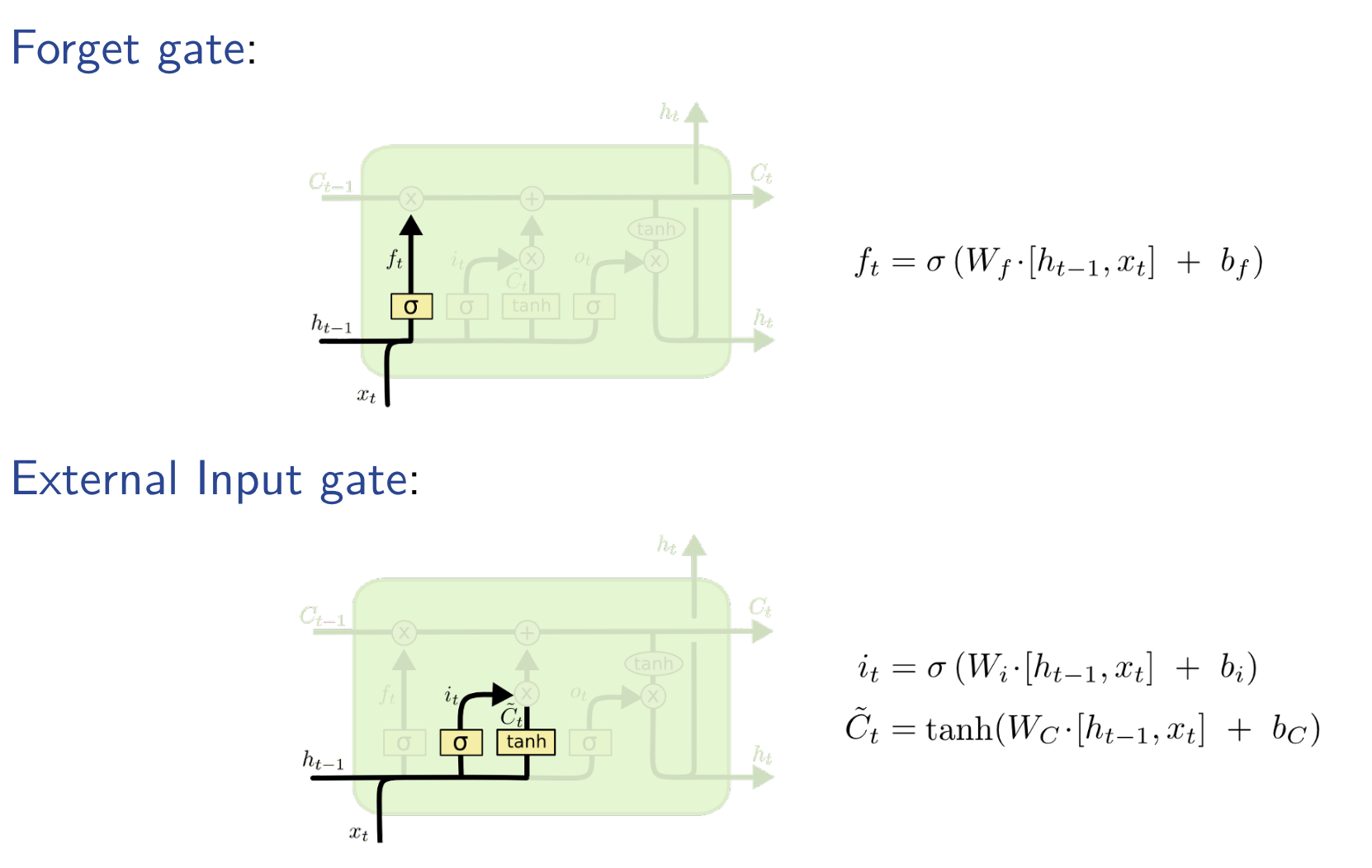
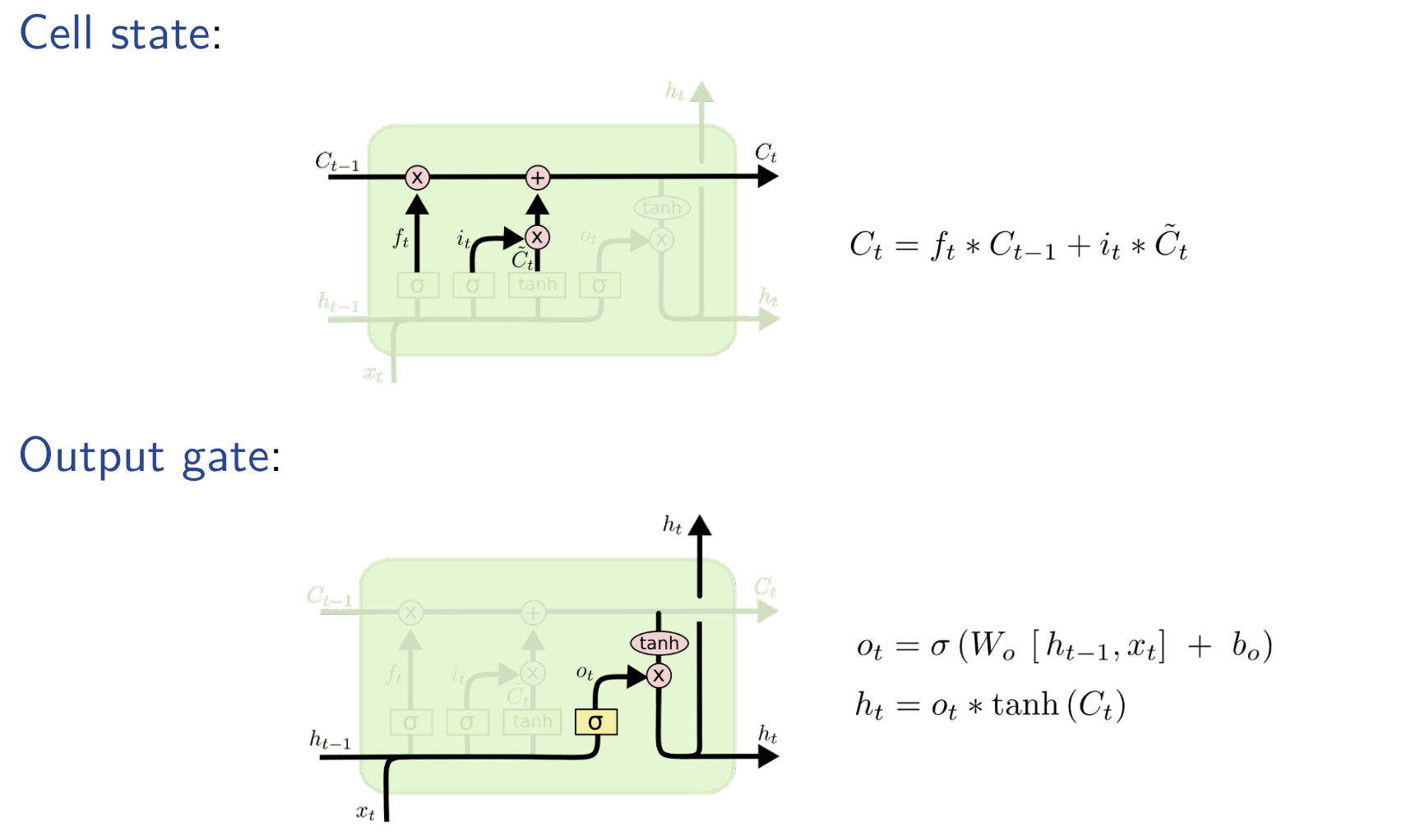
Total RNN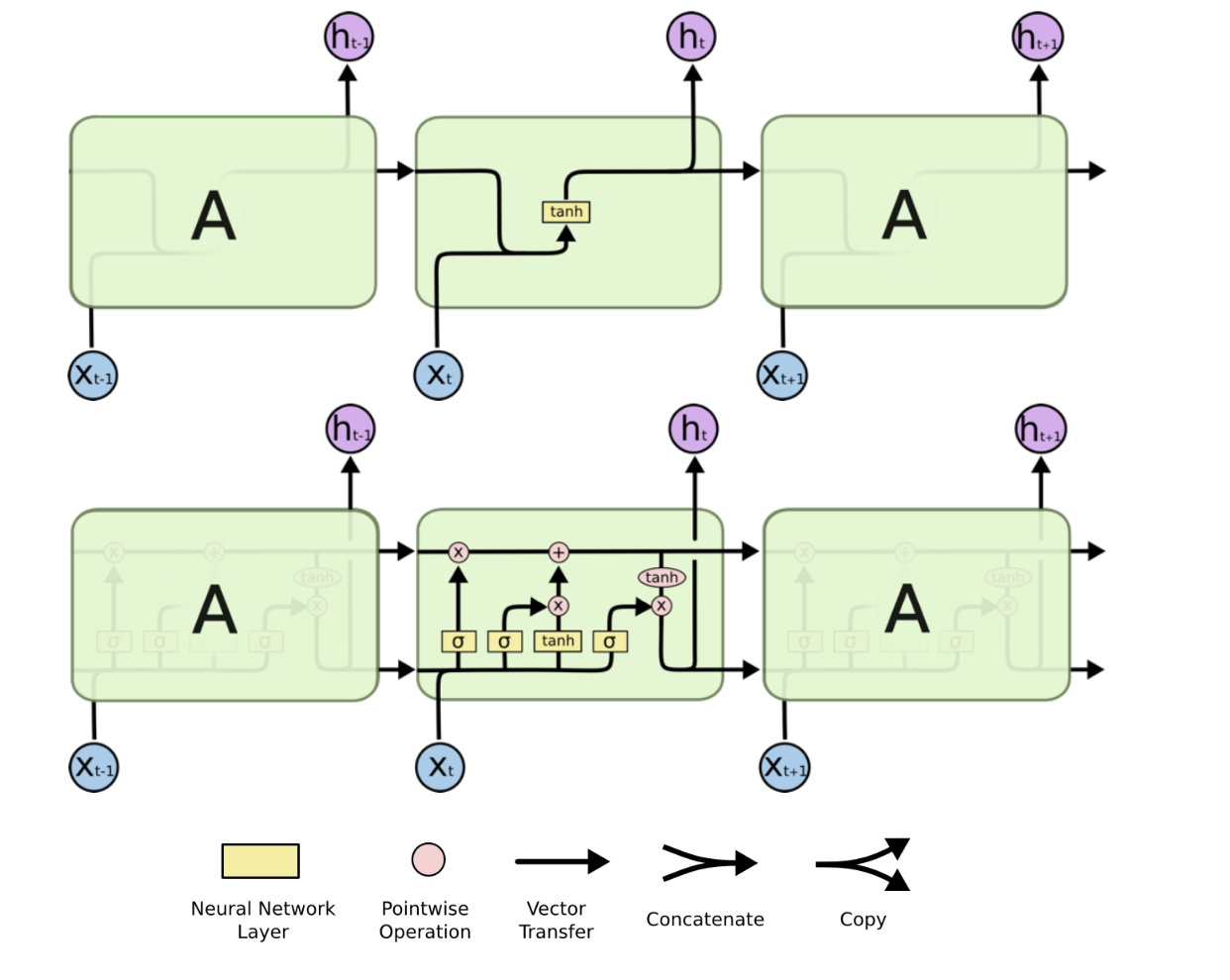
Ensemble Learning
Voting
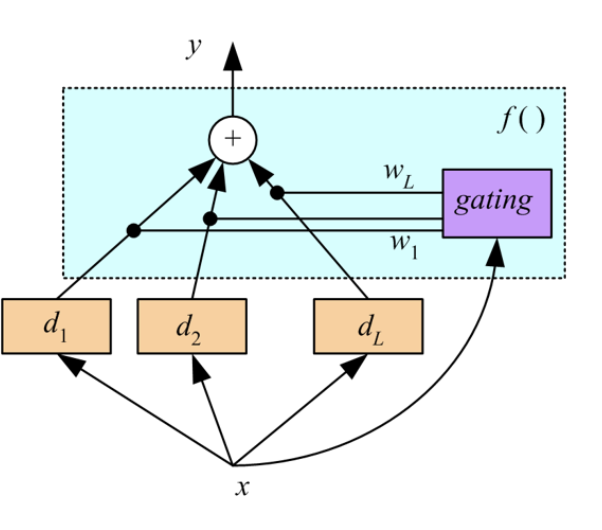
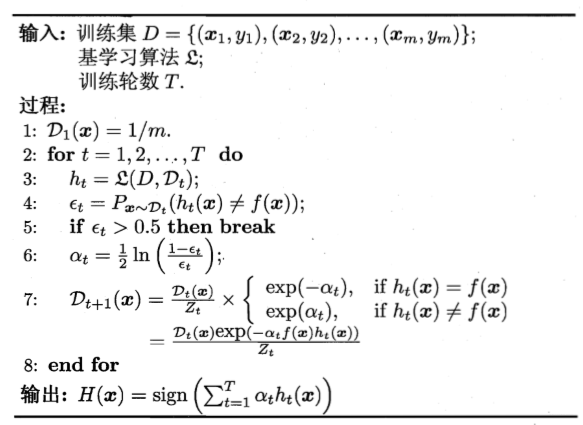
Boosting
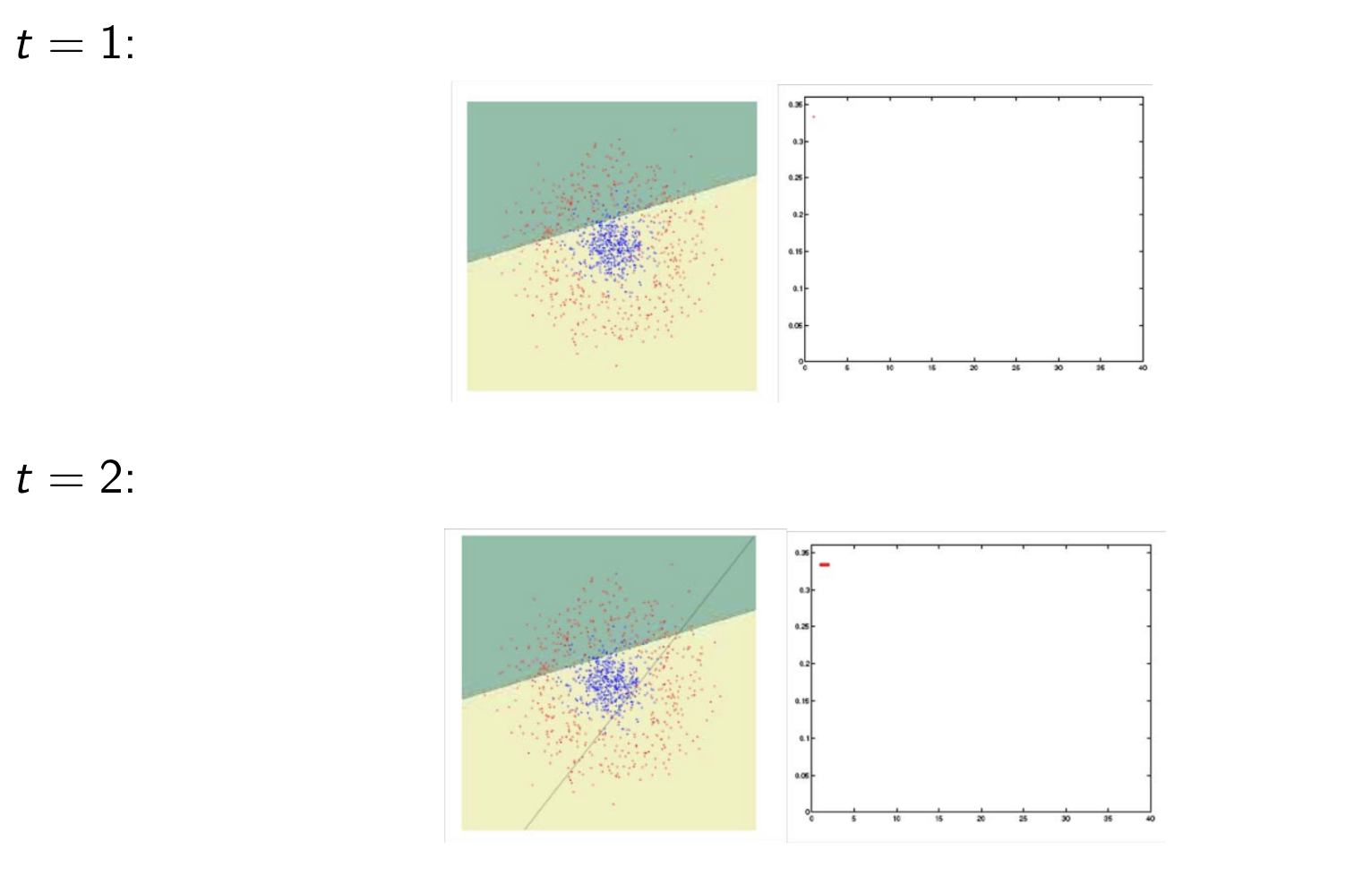
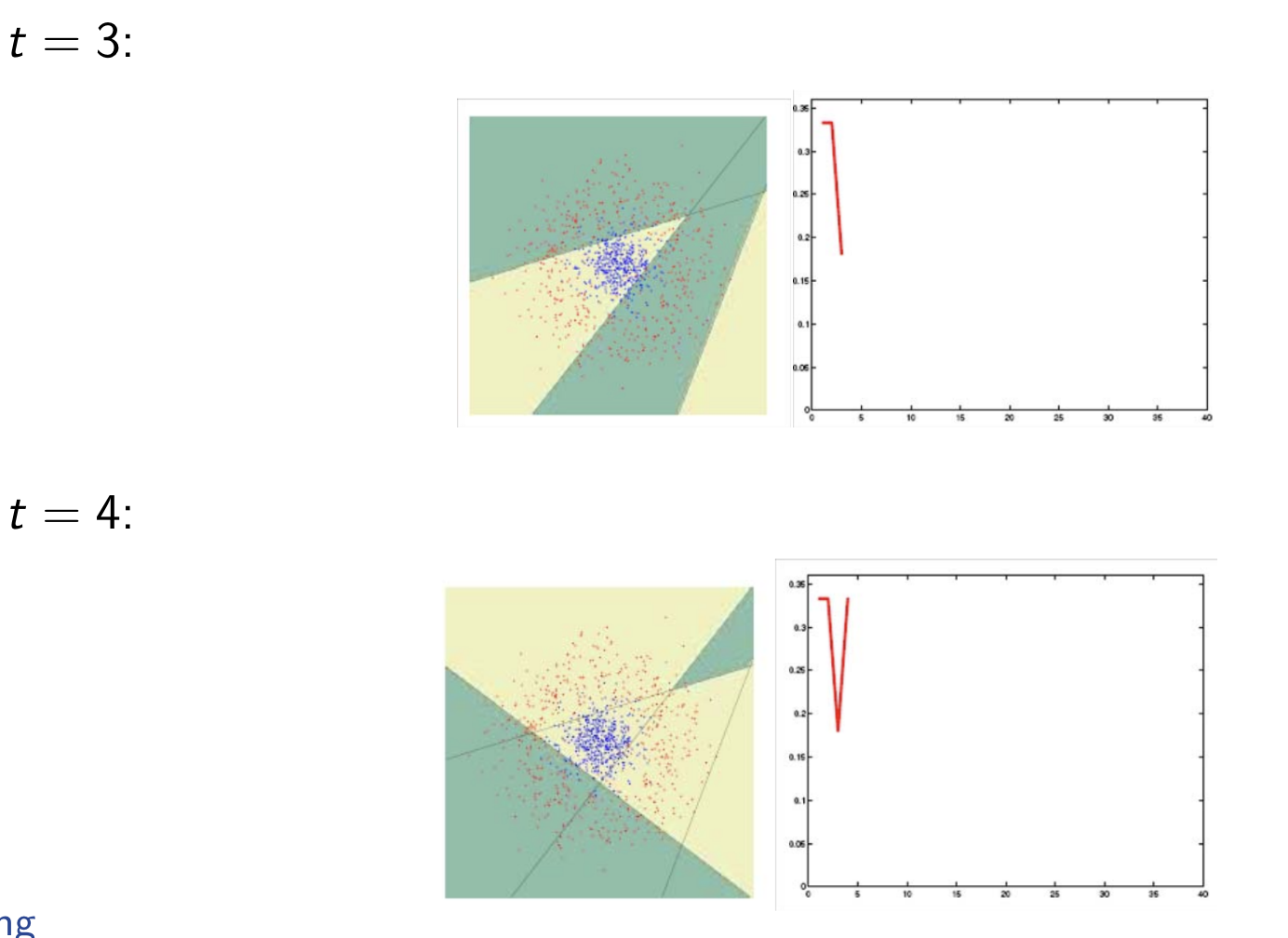
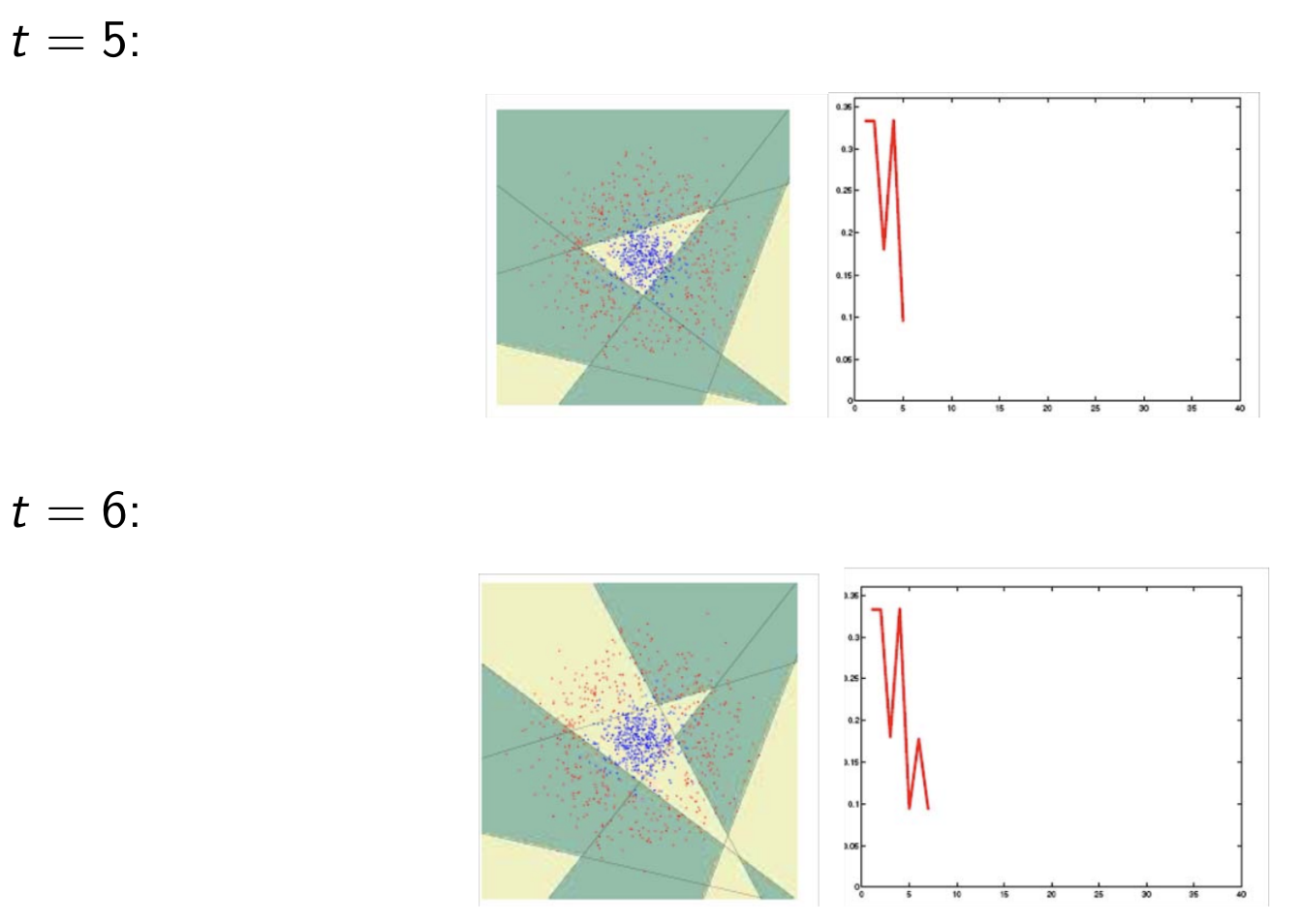
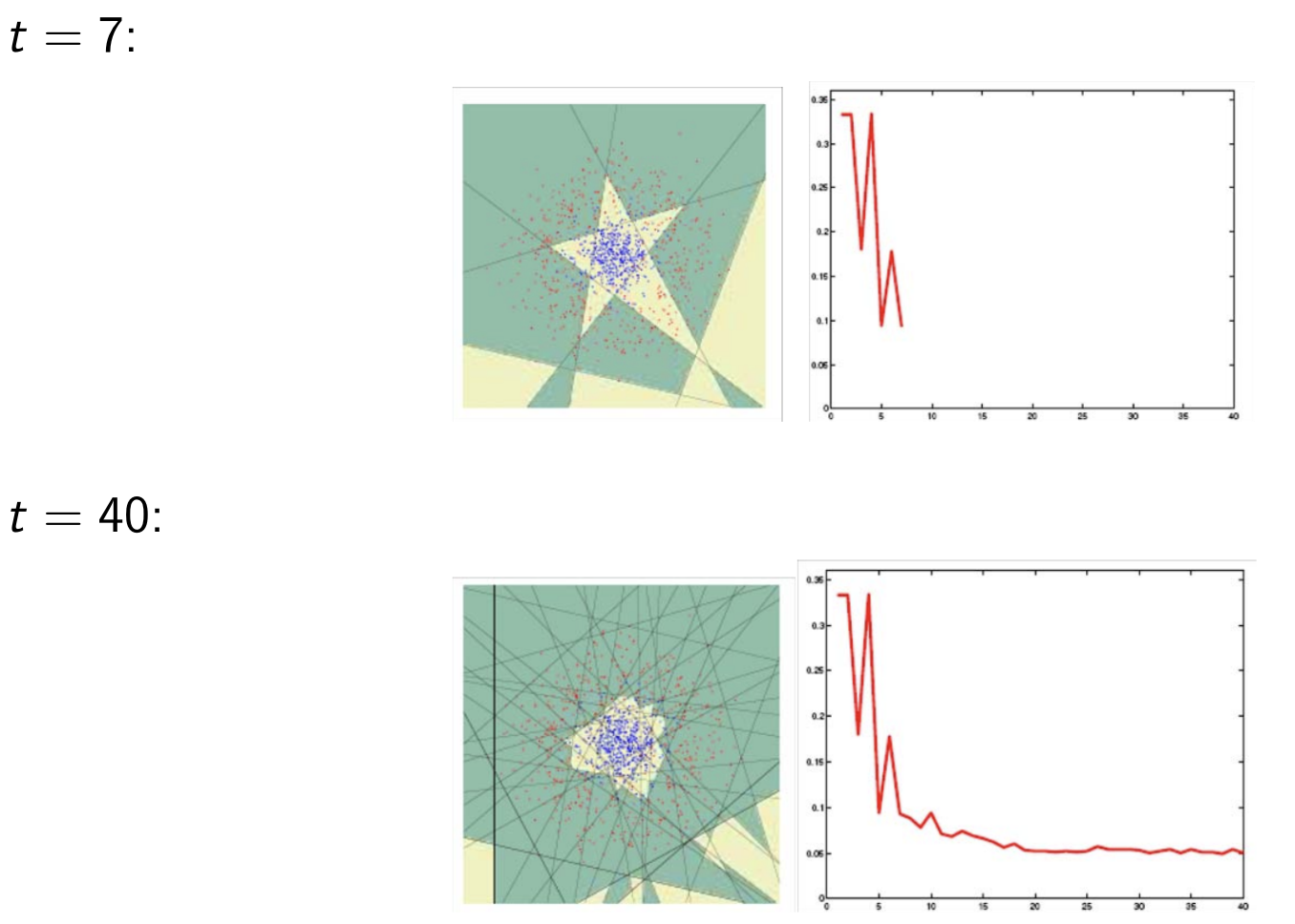
Model Assessment Selection
Cross-Validation and Resampling
K-Fold Cross Validation
The dataset
$$
\begin{aligned}
\mathcal{T}_1 &= \mathcal{X}_2 \cup \mathcal{X}_3 \cup \cdots \cup \mathcal{X}_K & \mathcal{V}_1 &= \mathcal{X}_1 \
\mathcal{T}_2 &= \mathcal{X}_1 \cup \mathcal{X}_3 \cup \cdots \cup \mathcal{X}_K & \mathcal{V}_2 &= \mathcal{X}_2 \
& \vdots & & \vdots \
\mathcal{T}_K &= \mathcal{X}_1 \cup \mathcal{X}2 \cup \cdots \cup \mathcal{X}{K-1} & \mathcal{V}_K &= \mathcal{X}_K \
\end{aligned}
$$
Any two training sets
5 × 2-Fold Cross-Validation
For each iteration, the dataset
$$
\begin{aligned}
\mathcal{T}_1 &= \mathcal{X}_1^{(1)} & \mathcal{V}_1 &= \mathcal{X}_1^{(2)} \
\mathcal{T}_2 &= \mathcal{X}_1^{(2)} & \mathcal{V}_2 &= \mathcal{X}_1^{(1)} \
\mathcal{T}_3 &= \mathcal{X}_2^{(1)} & \mathcal{V}_3 &= \mathcal{X}_2^{(2)} \
\mathcal{T}_4 &= \mathcal{X}_2^{(2)} & \mathcal{V}_4 &= \mathcal{X}_2^{(1)} \
& \vdots & & \vdots \
\mathcal{T}_9 &= \mathcal{X}_5^{(1)} & \mathcal{V}_9 &= \mathcal{X}5^{(2)} \
\mathcal{T}{10} &= \mathcal{X}5^{(2)} & \mathcal{V}{10} &= \mathcal{X}_5^{(1)}
\end{aligned}
$$
In total, we have done 5 times of 2-fold cross-validations.
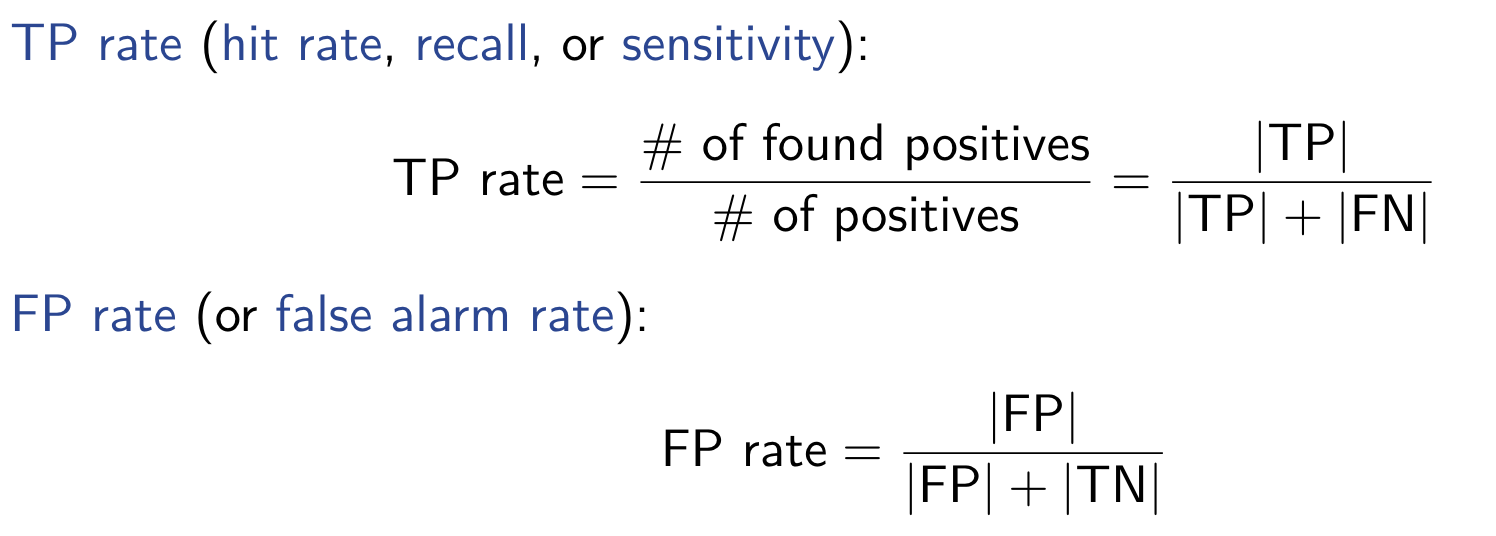
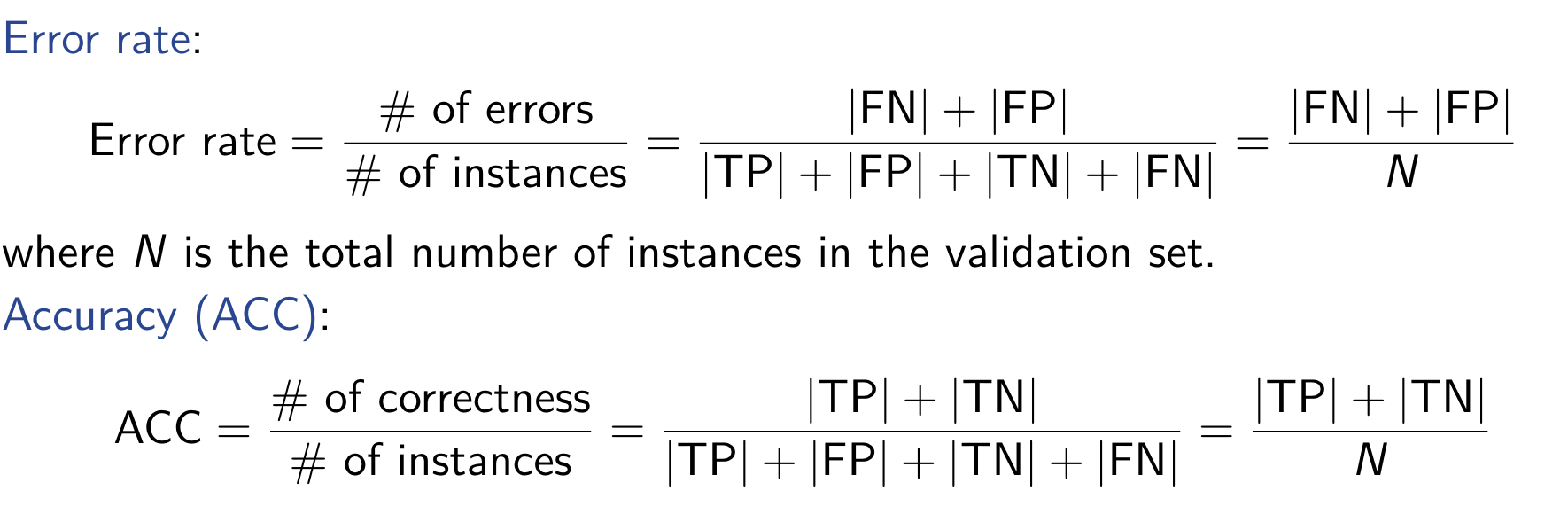
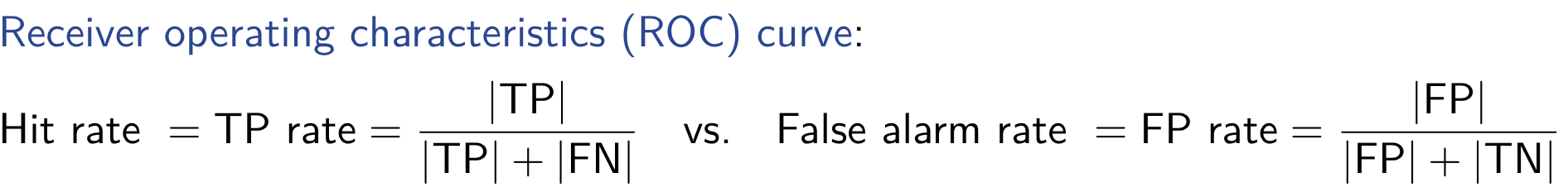
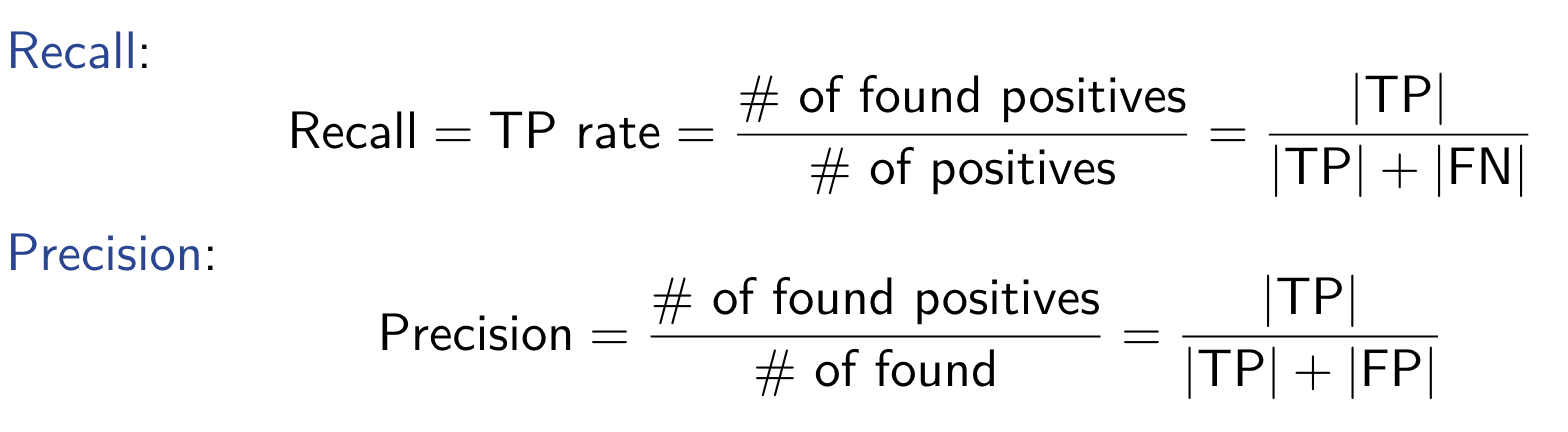
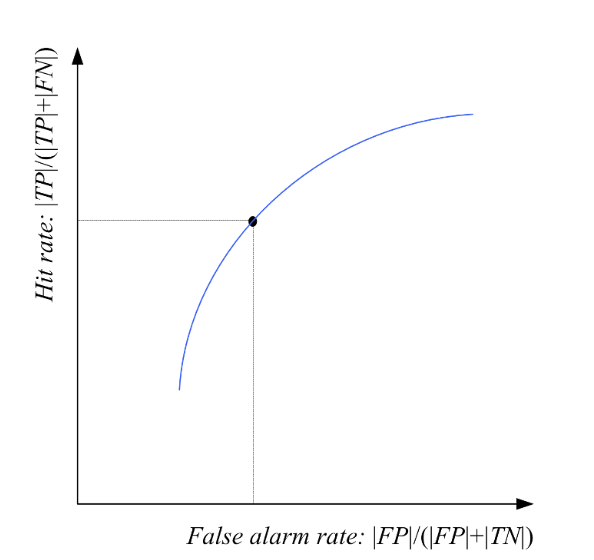
Interval Estimation
Two-sided confidence interval
or
One-sided confidence interval
and
Students’ t-distriburion
In the previous intervals, we used (\sigma); that is, we assumed the variance is known. When the variance (\sigma^2) is not known, it can be replaced by the sample variance
The statistic
For any
or
Hypothesis Testing
We can also define a one-sided test or one-tailed test.
Null and alternative hypotheses:
as opposed to the two-sided test when the alternative hypothesis is (\mu \neq \mu_0).
The one-sided test with level of significance (\alpha) defines a 100(1 - (\alpha)) confidence interval bounded on one side in which (m) should lie for the hypothesis not to be rejected.
We fail to reject (H_0) with level of significance
Otherwise, we reject
If the variance
E.g., we can define a two-sided t test :
We fail to reject at significance level
One-sided t test can be defined similarly.
Performance Evaluation
Test statistic:
where
We can define a one-sided t test :
We fail to reject at significance level
Performance Comparation
K-Fold Cross-Validated Paired t Test
Matrix Factorization
Non-negative Matrix Factorization
- Title: (CS182)Machine Learning Review
- Author: Gavin0576
- Created at : 2024-06-04 15:43:58
- Updated at : 2024-06-15 15:58:41
- Link: https://jiangpf2022.github.io/2024/06/04/Machine-Learning-Review/
- License: This work is licensed under CC BY-NC-SA 4.0.